Connected-Pathology and KU Leuven Institute for Single Cell OMICS (LISCO) announce a partnership to offer multiplex immunohistochemistry services
For decades pathologists have relied on making histopathological assessments based on the widely well-known principles of immunohistochemistry (IHC). Although conventional IHC is a powerful tool to assess disease pathophysiology, it remains greatly limited by the amount of biological information that can be extracted. However, histological sections, often being precious limited tissue sample material, offer highly specific disease-relevant phenotypic information, going beyond the usual single-marker analysis. With novel techniques on the rise, such as multiplexed IHC, it is now possible to perform high-throughput histopathological profiling within a single tissue section.
At Connected-Pathology, we revolutionize the future of digital histopathology services by providing our clients and partners with novel, cutting-edge techniques, helping them accelerate their research. We are happy to announce our partnership with the KU Leuven Institute for Single-Cell Omics (LISCO) to offer their proprietary MILAN platform as part of our digital histopathology service offering.
What is MILAN?
MILAN stands for Multiple Iterative Labelling by Antibody Neodeposition. MILAN leverages the principles of conventional fluorescent IHC together with controlled antigen retrieval and retention steps, resulting in multiplexed antibody labeling. Simply put, your sample will be stained using an optimized immunofluorescent (IF) protocol and subsequent high-resolution images will be recorded in their respective channels. Following the recording, antibodies will be removed while carefully preserving tissue antigens, whereby another staining round can begin. In this way, your regular fluorescent IHC assessments are taken to the next level by allowing up to 6 rounds of IF staining, resulting in up to 80 markers analyzed at the cellular level on a single tissue section.
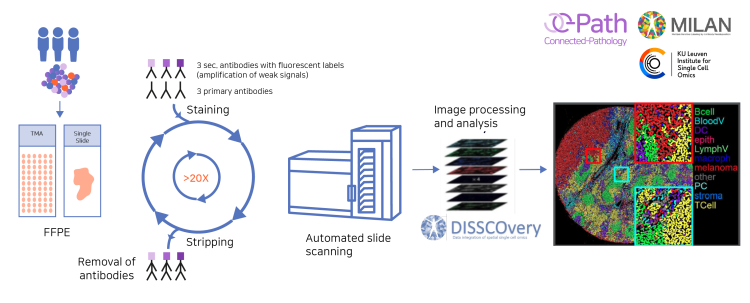
The MILAN workflow. Starting materials are formalin-fixed paraffin-embedded (FFPE) single-slide tissue sections or tissue microarrays (TMAs). One round of staining can be performed with up to 3 primary antibodies followed by 3 secondary fluorescently-labeled antibodies. Following labeling, high-resolution images are acquired in each fluorescent channel and the antibodies are stripped during the antigen retrieval protocol. Once all rounds are completed, all collected images are precisely superposed to obtain a multi-layer image containing all the different markers before performing the extended data analysis.
Why use MILAN?
MILAN is based on conventional fluorescence IHC principles, increasing the reliability and reproducibility of your results.
MILAN is a robust and versatile multiplex IHC method, allowing for whole-slide assessments. Equally, MILAN can be scaled down to the generation of tissue microarrays (TMA), enabling analysis of large cohorts in an efficient and affordable way.
No downtime when starting a project: MILAN is easily and rapidly integrated into your project workflow, as both primary and secondary fluorescent-labelled antibodies are commercially available, most of which are validated as part of the MILAN antibody library.
In case a non-validated antibody is requested, extensive antibody validation experiments are performed, producing significant time and resource savings by avoiding tedious and often frustrating antibody validation experiments.
Effective and proficient approach to extensive immunophenotyping studies by allowing the simultaneous visualization of both specific cell types and signaling proteins in the native tissue microenvironment.
Select from a wide range of disease-specific markers or a cell subtype-based panel.
Extensive immunophenotyping analyses will help you gain confidence in your candidate drug’s mode of action.
