New research discovers how plant hormones called brassinosteroids affect gene expression and plant growth
Hormones play an important role in the development of plants. Scientists have long known that networks of genes control cell development, but it has been difficult to untangle the complex relationships between hormones, cell identity, and developmental stage. Now, the team of Eugenia Russinova (VIB-UGent Center for Plant Systems Biology), together with international colleagues from Philip Benfey’s lab at Duke University (US), Iowa State University (US), University of British Columbia (Canada), and Humboldt Universitat zu Berlin (Germany), made great strides in understanding the relationship between plant hormones and gene regulatory networks. Their work appears in Science.
Brassinosteroids change gene expression
Understanding how plant hormones regulate gene networks in different cell types and at specific developmental stages could lead to improved plant growth in changing environments. In particular, hormones like brassinosteroids, auxin, gibberellins, and abscisic acid have unique effects on different tissues. However, it is still unclear how these hormone-induced gene networks are controlled in specific cells and at different stages of development.
Thanks to new technology like single-cell RNA sequencing and tissue-specific gene manipulation, researchers from the team of Eugenia Russinova (VIB-UGent Center for Plant Systems Biology), together with international colleagues from Philip Benfey’s lab at Duke University (US), Iowa State University (US), University of British Columbia (Canada), and Humboldt Universitat zu Berlin (Germany), made great strides in understanding the relationship between plant hormones and gene regulatory networks. They used the Arabidopsis root as a model system to study the effects of brassinosteroids, a group of plant steroid hormones that impact cell division and elongation during root growth.
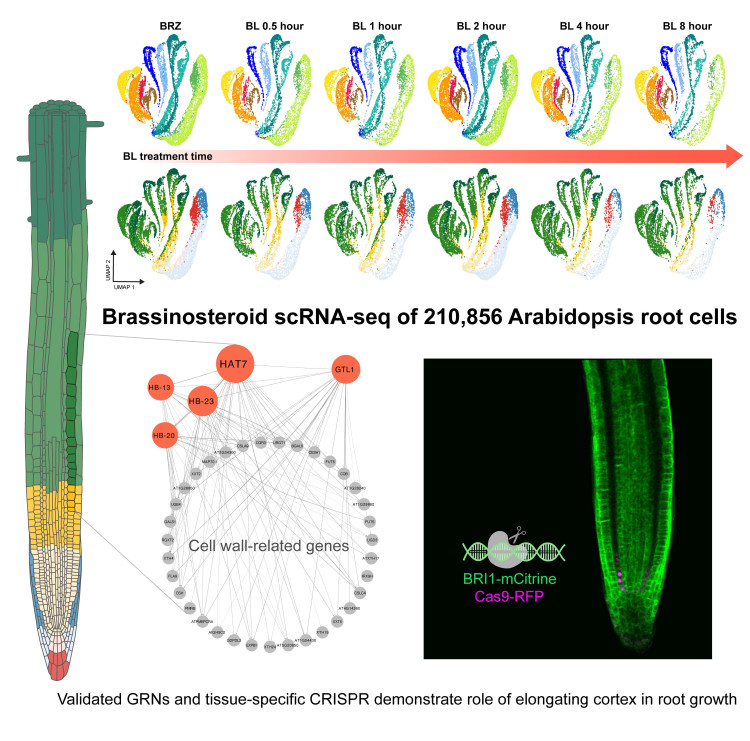
Dr. Trevor Nolan (Duke University), co-first author of the study: "We found that brassinosteroids affect gene expression in the elongating cortex, triggering a shift from proliferation to elongation that is associated with the upregulation of cell wall-related genes. In other words, brassinosteroids change gene expression, which leads to elongation of root cells."
Dr. Nemanja Vukašinović (VIB-UGent), co-first author of the study: “We validated single-cell RNA sequencing findings by inducing the loss of brassinosteroid signaling in the cortex using tissue-specific CRISPR, which lead to reduced cell elongation.
Finding regulators
Using time-course data, the researchers were able to propose brassinosteroid-responsive gene regulatory networks, which led to the identification of HAT7 and GTL1 as validated regulators of brassinosteroid response in the elongating cortex. The scientists analyzed over 210,000 single-cell transcriptomes, providing a detailed and high-resolution view of the brassinosteroid-mediated gene regulatory networks.
Prof. Jenny Russinova (VIB-UGent): "These findings demonstrate the power of single-cell genomics in identifying specific regulators of plant growth and development. By better understanding how gene networks are regulated in specific tissues and at specific developmental stages, we may be able to engineer plants that respond more effectively to changing environments and stress."
This research represents an important step forward in our understanding of how hormones control gene expression during cell development and could have important implications for agriculture and other fields.
The data from this study are publicly available and can be explored through an interactive browser.
Publication
Brassinosteroid gene regulatory networks at cellular resolution in the Arabidopsis root. Nolan, Vukašinović, Hsu, et al. Science, 2023.
